Photo courtesy of Philips Healthcare
Hollywood has shone its lights on potential dystopian futures and the threats of cruel, calculating computers for decades. But look beyond the Arnold Schwarzenegger-type cinematic portrayals of cybernetic antagonists, and there might be a future for the United Kingdom’s National Health Service (NHS) where teaching machines new tricks with data could result in huge benefits for transforming diagnostic capacity and the very nature of how our professionals identify serious illnesses.
Artificial intelligence is serious business, a point reinforced as recently as October when Cambridge University opened the doors to its new institute dedicated solely to the discipline. Technology giants like Google, IBM Watson and Dell EMC are also throwing their weight into the field.
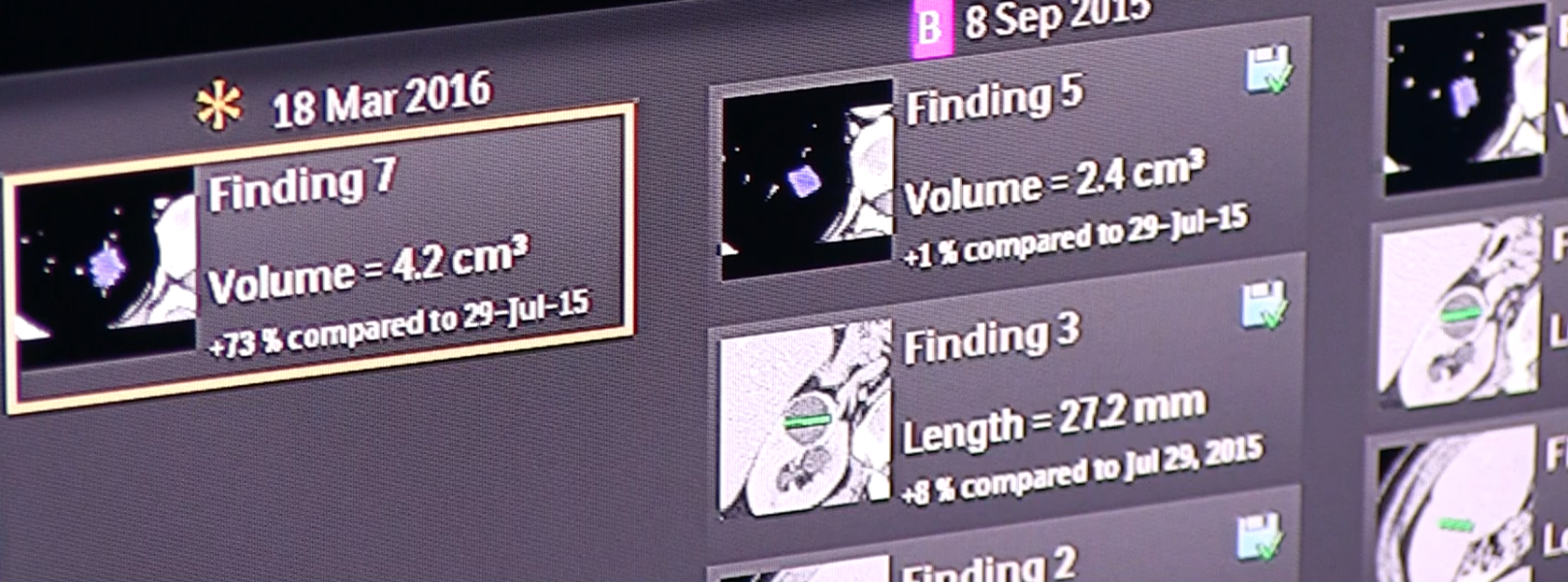
But there is one specific area where machine learning could unleash one of the NHS’ biggest untapped resources — a huge archive of diagnostic imaging. In radiology alone, millions of X-rays, computed tomography (CT) scans and a plethora of other diagnostic imaging have been stored digitally for many years, for the most part sitting in departmental archives undisturbed.
This vast data source is only set to grow as other “ologies” look to invest in extensive digitization programs. Pathology, for example, has already digitized in other parts of the world, with pioneering NHS trusts starting to take the lead in shifting the U.K.’s cellular examinations from cumbersome slides to high-resolution digital images that can be quickly shared and analyzed across professional and organizational boundaries.
Rich sets of data from these disciplines and the full mix of other diagnostic departments may very soon be accessible from a single storage point as hospitals look to consolidate imaging from a multitude of specialties into central VNAs which, if effectively deployed, will allow enterprise level access to a comprehensive and powerful array of patient images.
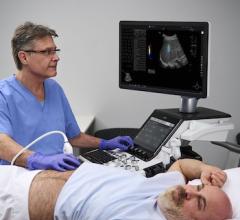
If this access can be opened to machine learning, the role of the archive will change from just being a big bucket that is occasionally accessed for retrieving data, to becoming a central component of the NHS enterprise, where data can be reprocessed and re-analyzed continuously to add clinical decision support for precision and personalized medicine based on population comparisons.
Machines to Proactively Diagnose Illnesses?
To put all of the above simply, the NHS has a wealth of imaging data that it can use to start teaching machines how to recognize parts of the human anatomy, and more importantly, how to recognize abnormalities.
The imaging archive has often been seen as a challenge, a nuisance and a necessary evil. Now our archives have potential to go from being largely unused storage boxes to goldmines of clinical knowledge.
Take the diagnosis of pneumonia as just one example. Once able to recognize the condition, machines could potentially search through imaging in advance of reports being carried out and flag it for the diagnostic specialist, or to the clinician, patients who appear to be at risk.
Machines can act as a powerful decision support system, but more than that, they could be programmed to proactively suggest other conditions that they have been taught to recognize. For example, whether the healthcare professional has considered that a patient might be suffering from lymphoma or asbestosis.
Getting the Right Technology in Place
All of this might sound a long way off, and there are certainly many cultural and technological hurdles to overcome, but the debate of whether a machine can play a much greater role in diagnostic reporting is an important one that raises many questions for the NHS. Can we increase the capacity of diagnostics to interrogate far more imaging and report on far more than possible with increasingly finite resources? Can our professionals, who are facing greater demand than ever, be given a much greater level of support? And can artificial intelligence help with the preventative, early intervention agenda now strongly being called for?
Leveraging data on a larger scale will obviously raise questions on patient integrity and anonymization — where there are challenges yet to be solved. Equally, we need to ask questions around data retention; do we purge data after the mandatory retention period that in the future could be used for data mining, comparisons and analysis? However we arrive at the answers, the opportunity from machine learning is too great to ignore, and we must lay the technological groundwork so that we have the capacity to make this a reality.
For machine learning to really work will take significant time, but we could get a head start today by utilizing a wealth of imaging already in existence, to begin to train technology to be able to identify patient conditions. The more data we feed into the system over time, the greater the accuracy of decision support will become.
To really make this work, technologies like the earlier mentioned VNA need to provide work on open architectures that are able to integrate and feed sources of data from modalities across healthcare into the research and wider analytical pool. We cannot look only to vendors to answer the challenges. Different applications will need to benefit from the underlying data to achieve the most for patients.
It is important to establish open platforms for efficient imaging data management. Openness is essential if we are to encourage cooperation amongst vendors and to stimulate market innovation.
Ultimately, this is about the patient. Today the follow-up of what actually happened to a patient following a series of activities is too rare. It takes time and resources and this is where IT is the enabler, allowing new technologies to discover relationships and risk factors and allowing this to be fed back into the clinical practices, and support clinicians in future decision making.
The eminent Prof. Stephen Hawking has said that artificial intelligence will be “either the best, or the worst thing, ever to happen to humanity.” For it to be the best for healthcare, we must embrace our data, but also be aware that the step from achieving results in research to valuable solutions in the frontline clinical practice will require a good deal of innovation, time, hard work and detailed understanding of workflows.
This is not about creating magic boxes that will solve everything sometime in the future, but about collaboration across the healthcare industry to enhance sustainability and deliver the best for patients.
Read the article “How Artificial Intelligence Will Change Medical Imaging.”
Watch the VIDEO “Machine Learning and the Future of Radiology.”
Chris Scarisbrick is sales director for Sectra U.K. and Ireland.


 March 06, 2026
March 06, 2026